


The full characterization of proteins in individual cells has often been overshadowed by transcriptomics. However, this is about to change.
On September 20th, 2021, the Technology Feature column of Nature magazine published an article titled "Single-cell proteomics takes center stage" [1]. According to the commentary, for a long time, transcriptomics has been the primary method of obtaining the complete protein composition of a single cell. However, the emergence of spatial single-cell proteomics technology is expected to bring significant changes and enable the study of the complete life cycle of single cells.

The advent of single-cell omics technologies has brought a revolution in biological research over the past decade. Despite this, research at the single-cell level has mainly focused on cell imaging and genome and transcriptome sequencing analysis, lacking the ability to perform in-depth characterization of high-value analysis of proteins, post-translational modifications, and proteome dynamics, which are more directly linked to phenotype. With the emergence of single-cell proteomics, the protein expression levels in individual cells have transitioned from being "inferred" based on mRNA abundance to being measured directly, thus representing a significant shift in the field.

Fig. A dream of single-cell proteomics [2]
The goal of single-cell proteomics is to classify and characterize the protein profile of a single or very small number of cells. Due to the non-amplification characteristics of proteins, high sensitivity quantitative techniques and instruments are required. Currently, scientists use antibodies or mass spectrometry to detect the proteome of individual cells. Antibodies are relatively simple to use and techniques such as flow cytometry or CODEXD can quantify up to 50 proteins in a single cell. For instance, Diana Mahdessian et al. identified 1180 proteins in a single U2OS cell for the first time by designing almost a thousand antibodies and found that approximately 539 of them were related to cell cycle function [3]. However, not all proteins have corresponding antibodies, and some antibodies only bind weakly or non-specifically to proteins. Furthermore, antibody-based methods only target specific proteins and can only observe part of the proteome, thus having a "preset boundary" for research. As a result, many scientists have turned to mass spectrometry-based approaches that enable the untargeted "full-discovery" of the proteome of individual cells.
Fig. Two paths to the proteome [4]
Mass spectrometry-based approaches have the ability to detect substances at the attomole (10-18 moles) level, making it sensitive enough to identify proteomes at the single-cell level. The latest 4D mass spectrometer has an ultra-high sensitivity, giving it a significant advantage in trace sample analysis. While single-cell proteome studies typically identify around 1000 proteins in a single cell, which is lower than the depth of transcriptomic analysis, it is still sufficient to reveal biological differences. For example, a recent study led by Matthias Mann's team used single-cell proteomics to identify the number of proteins in different stages of the cell cycle, ranging from 611 in the growth phase of cell division to 1263 in the subsequent phase of DNA synthesis. The results revealed key regulators of the cell cycle, such as CDK2NA, E2 ubiquitin ligase UBE2S, DNA topoisomerase TOP2A/B and chromatin regulator HMGA1. [5]

Fig. T-SCP correctly quantifies cell cycle states [5]
The use of large-scale single-cell proteomics analysis has provided valuable insights into cellular heterogeneity in both healthy and diseased states. In a recent study published in Nature Communications in July 2021, the team led by Erwin M. Schoof from the University of Copenhagen utilized mass spectrometry-based single-cell proteomic technology to quantify approximately 1,000 proteins in a single cell from a primary leukemia model system. This study shed light on the cellular heterogeneity present in the progression of leukemia, offering a valuable contribution to our understanding of this disease[6].

Fig. Experimental overview of workflow [7]
It is important to note that traditional single-cell experiments do not provide information on the location of cells in the tissue, despite the fact that cell location within the tissue can be crucial to understanding biological processes. The latest proteomic research has overcome this limitation by incorporating spatial dimension information, resulting in the emergence of "spatial proteomics". Researchers Ryan Kelly and Ying Zhu from the Pacific Northwest National Laboratory have used a combination of nanoPOTS technology for trace sample processing, laser capture microdissection (which uses a laser to remove cells from tissue), and mass spectrometry to generate quantitative, cell type-specific images of over 2000 proteins at 100-µm spatial resolution on tissue sections of the mouse uterus prepared for blastocyst implantation [7]. This approach provides a beautiful portrait of the spatial heterogeneity of the proteome in tissues.
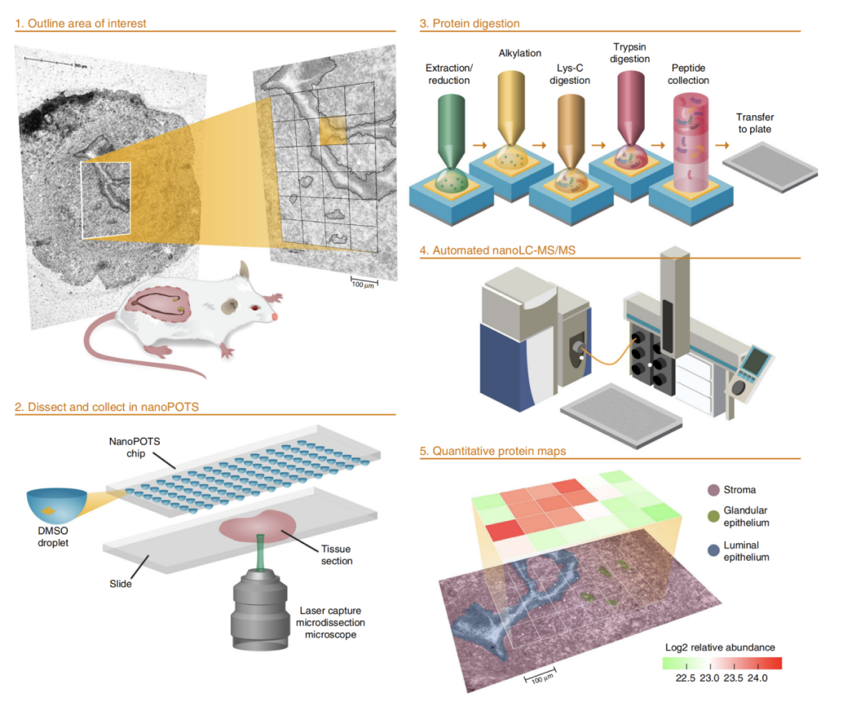
Fig. Schematic workflow for high-throughput, spatially resolved proteomics using the nanoPOTS imaging platform [7]
Single-cell proteomics has become a highly sought-after research area in proteomic technology, and has recently received a significant boost in funding from the US National Institutes of Health, with approximately $20 million being allocated towards the technical development of single-molecule and single-cell proteomics. This highlights the international attention and emphasis being placed on single-cell proteomics research. In the domestic market, PTM BIO, a leading company in the field of proteomics, has launched their "true single-cell proteomics" analysis, enabling the full discovery and analysis of proteomes down to a single cell. This breakthrough technology is expected to facilitate new discoveries and advancements in the fields of life science and medicine.
For more details, please call the hotline (400-100-1145) or consult local sales engineers.
Reference
1. Jeffrey M. Perkel, 2021, Single-cell proteomics takes centre stage. Nature.
2.Vivien Marx, 2019, A dream of single-cell proteomics. Nature.
3.Diana Mahdessian, et al, 2021,Spatiotemporal dissection of the cell cycle with single-cell proteogenomics. Nature.
4.Nikolai Slavov, 2020, Unpicking the proteome in single cells. Science.
5.Brunner, A.-D. et al, 2021, Ultra-high sensitivity mass spectrometry quantifies singlecell proteome changes upon perturbation. Preprint at bioRxiv.
6.Erwin M. Schoof, et al., 2021, Quantitative single-cell proteomics as a tool to characterize cellular hierarchies. Nature Communications.
7.Paul D Piehowski , et al., 2021, Automated mass spectrometry imaging of over 2000 proteins from tissue sections at 100-μm spatial resolution. Nature Communications.