



A dream of single-cell proteomics [1]
Introduction
Single-cell proteomics is the "crown jewel" in the development of proteomics technology, and is a dream pursued by scientists [1]. With the advent of the post-genome era and the development of sequencing technology, proteomic analysis at the single-cell level provides the most essential information for the study of cellular heterogeneity in biological systems, the construction of fine protein maps, and the analysis of spatial protein networks. Moreover, the accurate information of proteomic analysis has indispensable application value in important fields such as tumor heterogeneity, stem cell differentiation, and germ cell development research.
Backgroud
Due to the limitation of extremely trace sample preparation, ultra-sensitive high-resolution biological mass spectrometer and data repeatability, single-cell proteomic analysis has been stuck in the stage of "dreams " for a long time. The single-cell proteomics analysis currently on the market is not a single-cell analysis in the true sense. It usually requires 103 cells, and the detection depth can only reach hundreds of proteins with poor data repeatability, making the current single-cell proteomic analysis does not yield truly valuable information. As a leading company in the field, PTM BIO has been committed to the continuous innovation of proteomics technology by virtue of its strong scientific and technological strength and tenacious spirit of exploration. Recently, PTM BIO has developed a product for true single-cell proteomic analysis, and launched the [True Single-cell Proteomics] analysis service to help science research in basic and translational medicine.
Advantages of true single-cell proteomics products
High accuracy and specificity
Accurate analysis of single cell proteomics
High detection depth and sensitivity
4000+ detection depth of a single type of trace cells
High repeatability and stability
The R index of the result correlation of three repeated samples can reach 0.935
Our internal data demonstrates that we have successfully achieved a detection depth of 1000+ proteins in a single cell (using HeLa cells as an example). At the 10-cell level, we have achieved a detection depth of 2000 proteins, and at the 100-cell level, we have detected 4000+ proteins (Figure 1-2). Repeated analysis of three proteomic datasets for 10 HeLa cells showed a coincidence degree of 70% for identification of the same protein, with a Pearson correlation coefficient R value of 0.935 (Figure 3). Quality control data confirmed extremely high accuracy and reliability of the analysis results, as peptide isotope peaks accurately and stably appeared in the spectrum during the quantitative analysis of single cells (Figure 4).
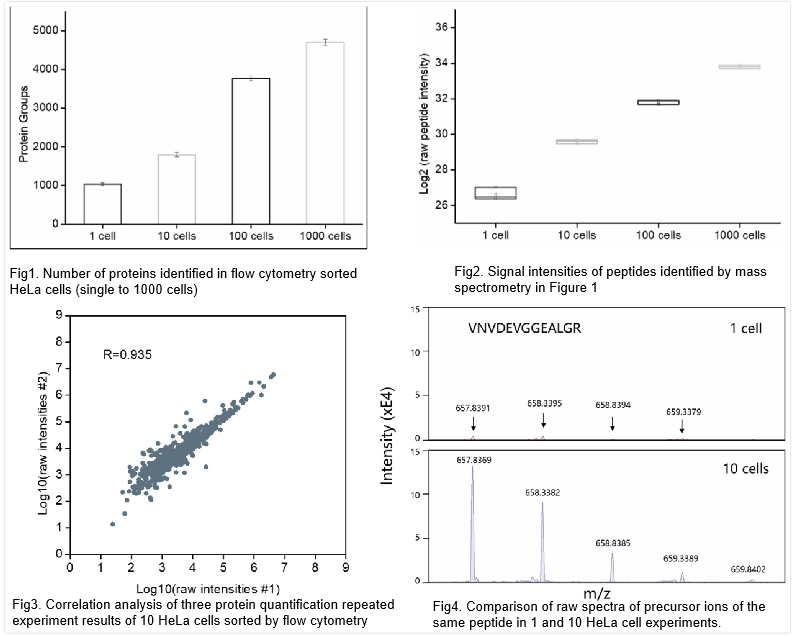
Figure 1. Number of proteins identified in flow cytometry sorted HeLa cells (single to 1000 cells). Figure 2. Signal intensities of peptides identified by mass spectrometry in Figure 1. Each cell amount was repeated three times. MaxQuant software was used for data analysis, and the FDR of PSMs and FDR of proteins were strictly limited to less than 1%. Figure 3. Correlation analysis of three protein quantification repeated experiment results of 10 HeLa cells sorted by flow cytometry. From flow cytometry sorted cells to sample preparation and mass spectrometry analysis, the three experimental processes are completely independent. And a scatter plot was made based on the logarithmic value of the original intensity of protein identification, which achieved good repeatability (Pearson correlation coefficient was 0.935). Figure 4. Comparison of raw spectra of precursor ions of the same peptide in 1 and 10 HeLa cell experiments. The peptide was identified in both 1 and 10 HeLa cell samples. From the comparison of the original spectrum, it can be seen that although the spectrum signal of the peptide in 1 cell is weak (about 10 times difference), it still has a high signal-to-noise ratio and a complete distribution of isotope peaks, proving the ultra-high sensitivity and accuracy of the single-cell proteomic platform.
True Single Cell Proteomics Product
Solutions in eight application areas
The analysis of true single-cell proteomics is revolutionary, as it allows for the analysis of extremely trace samples (such as stem cells, germ cells, embryonic cells, nerve cells, etc.) [2-4]. This breakthrough will undoubtedly lead to new discoveries and advancements in the following areas:
1. Develop a precise organ proteome map
Organs are composed of various types of cells, and studying the proteome of each cell type at the single-cell level can provide a deeper understanding of the mechanisms that regulate organ function and the pathogenesis and progression of degenerative diseases affecting organs.
2. Exploring tumor heterogeneity and investigating drug resistance mechanisms
Tumor tissue is highly heterogeneous, with cells continuously clonally evolving to become more aggressive through spontaneous mutations. Under the pressure of chemotherapy and targeted drugs, these mutations can result in the acquisition of drug resistance by some tumor cells. Therefore, analyzing the proteome of tumor cells at the single-cell level can help establish a more precise relationship between gene mutations and protein expression, enabling us to infer different mechanisms of drug resistance. This information provides valuable theoretical support for subsequent drug compatibility and the development of new anticancer drugs.
3. Analysis of circulating tumor cells
Liquid biopsy targeting circulating tumor cells (CTCs) is a crucial breakthrough in precision medicine for cancer research. Single-cell proteomic analysis of CTCs provides critical protein-level data, revealing crucial information such as the malignancy level of tumor tissue and the activation status of signaling pathways, which can help to develop more precise treatment plans.
4.Research on immune system and tumor immunity
Single-cell proteomic analysis of immune cells is crucial for understanding the intricate differentiation and regulation of various immune cell types and their role in maintaining immune homeostasis. Moreover, such analysis can shed light on the mechanisms underlying immune-related diseases. Additionally, gaining a comprehensive understanding of the protein expression profiles of different tumor-infiltrating lymphocytes (TILs) can facilitate the design and development of next-generation tumor immunotherapy drugs.
5. Analysis of cell therapy
Cell therapy, specifically CAR-T, has seen rapid development in recent years. However, effective methods for analyzing how CAR-T cells enter the tumor microenvironment are still lacking. Single-cell proteomics analysis can provide information on the activation state of CAR-T cells, as well as changes in the CAR-T proteome at different time points, assisting drug development scientists in the design and optimization of chimeric antigen receptors (CAR).
6. Neuroscience research
Single-cell proteomic analysis is a powerful tool for studying the cellular makeup of the mammalian nervous system, which is highly heterogeneous and complex. By analyzing the protein expression patterns of individual neural cells, researchers can gain a more detailed view of neural development and differentiation, as well as the cellular and molecular mechanisms of normal brain function and disease.
7. Reproductive studies
Single-cell proteomics has revolutionized the study of germ cells such as oocytes and spermatogonia, which are often limited in number during proliferation and differentiation. Traditional proteomics analysis is limited by the amount of available material, but single-cell proteomics can analyze the proteome of individual cells, allowing for more detailed and accurate information on the protein expression patterns of germ cells.
8. High throughput drug screening
Single-cell proteomic analysis is a promising approach for high-throughput drug screening using human primary cells. It allows for the observation of drug efficacy and potential toxicity on cells with different functions and provides a deeper understanding of the mechanism of drug action. This breakthrough in sample size limitation can greatly improve the success rate of drug development.
For more details, please call the hotline (400-100-1145) or consult the local PTM BIO sales engineers.
Reference:
1. Vivien Marx, 2019, A dream of single-cell proteomics.Nature Methods.
2. Mahdessian, D., et al., 2021, Spatiotemporal dissection of the cell cycle with single-cell proteogenomics.Nature.
3. Mahmoud Labib., 2021, Single-cell analysis targeting the proteome.Nature Reviews Chemistry.
4. Z. Bai, et al, 2021, Single-cell Analysis Technologies for Immuno-oncology Research: from Mechanistic Delineation to Biomarker Discovery.Genomics, Proteomics & Bioinformatics.