Service Introduction
PTM BIO has launched a new generation of 4D-DIA proteomics technology applications that combine 4D proteomics with data-independent acquisition (DIA), based on the revolutionary DIA PASEF technology developed in collaboration with research teams such as Matthias Mann and Ruedi Aebersold. By combining the strengths of 4D instrument and DIA technology, 4D-DIA proteomics represents a major upgrade to proteomics technology and overcomes some of its original limitations. This innovative technology is particularly suitable for proteomics scientific research on large-scale samples, such as clinical cohort samples and crop population traits.
Features
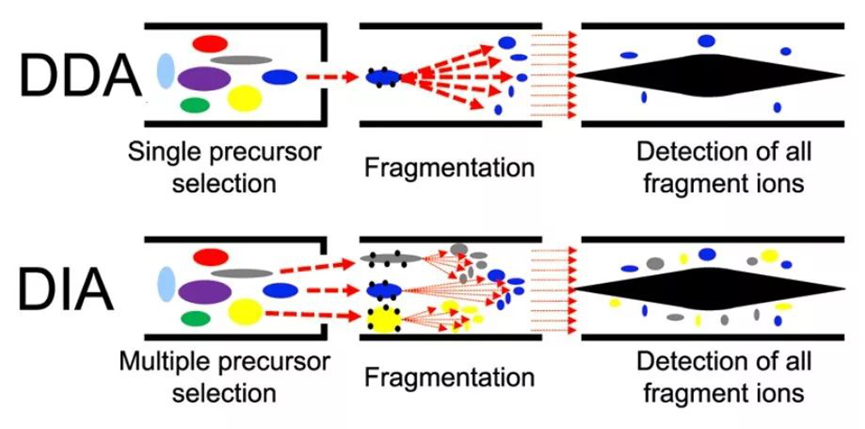
Proteomics techniques can be classified into two main strategies: data-dependent acquisition (DDA) and data-independent acquisition (DIA).
1. DDA strategy: Collect strong and high-abundance peptide ions within each time window during primary mass spectrometry, which are then subjected to fragmentation analysis during secondary mass spectrometry;
2. DIA strategy: Divide the entire full scanning range of mass spectrometry into several windows, and cycle all ions in each window for selection, fragmentation, and detection.
The DIA technology can obtain all fragment information of all ions in the sample without any omission. However, its data acquisition strategy may result in a highly complex spectrogram, posing significant challenges for data analysis. Additionally, its quantitative reliability is still a matter of controversy.
The new generation technology, 4D-DIA, represents a comprehensive upgrade of the DIA strategy. By innovatively combining the advantages of 4D proteomics and DIA technology, it not only maintains the excellent quantitative parallelism of DIA technology but also solves the issue of detection reliability to a great extent. Furthermore, it also enhances protein recognition ability, detection sensitivity, and data integrity.
Technical Advantages
1. Nearly 100% ion utilization greatly improves detection sensitivity
In the diaPASEF scanning mode, there is a correlation between the CCS value and m/z associated with ion mobility. Therefore, the four-stage rod can utilize this feature to gradually scan and acquire nearly 100% of the ion signal, significantly enhancing the sensitivity and depth of detection. In contrast, only a small part of these signals can be collected in DDA and traditional DIA modes.
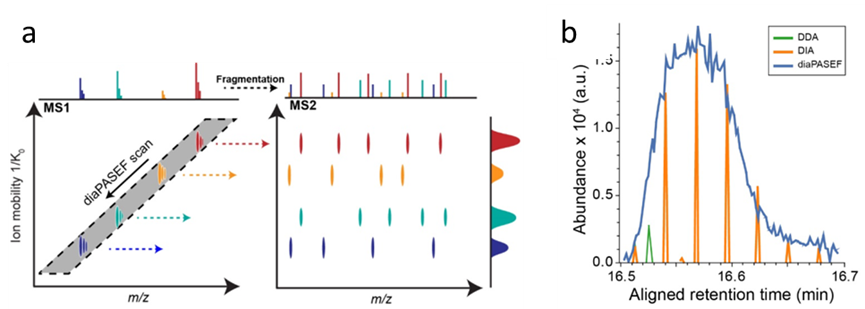
2. Significantly increased indentification depth
The 4D-DIA technique significantly enhances the depth of proteomic detection, enabling the identification of over 7500 proteins in a single 120-minute experiment with 200ng of HeLa lysate. Moreover, three experiments have collectively quantified 6974 proteins with data integrity up to 96%. In contrast, traditional DIA technology typically requires a μg-sized sample to detect approximately 5000 proteins.
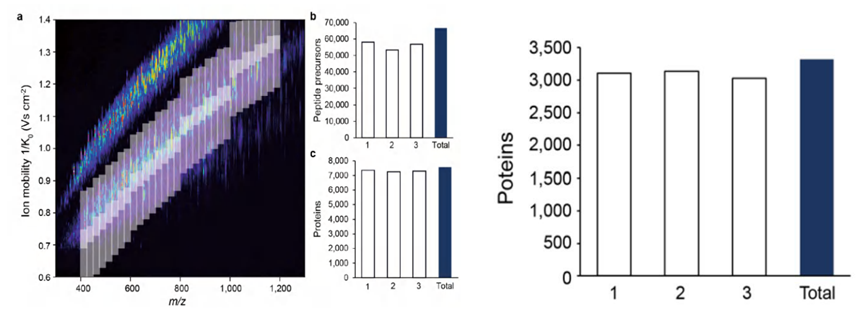
3. Comprehensively improve quantitative integrity
With superior instrument performance and advanced acquisition methods, 4D-DIA technology can push the limits of sensitivity even further. For example, in a 120-minute single-needle experiment with just 10 ng of HeLa lysate, 3323 proteins were identified using 4D-DIA technology, compared to 2723 proteins in the 4D-DDA pattern under the same experimental conditions. Even in such trace cases, 4D-DIA achieves 85% data integrity in protein quantification.
Reference
-
Yang QS, Wu JH, et al. Quantitative proteomic analysis reveals that antioxidation mechanisms contribute to cold tolerance in plantain (Musa paradisiacal L.; ABB Group) seedlings. Mol Cell Proteomics. 2012; 11(12): 1853-69.
-
Leivonen SK, Rokka A, et al. Identification of miR-193b targets in breast cancer cell and systems biological analysis of their functional impact. Mol Cell Proteomics.2011; 10(7).
-
Xu C, Gao X, et al. The basal level ethylene response is important to the wall and endomembrane structure in the hypocotyl cells of etiolated arabidopsis seedlings. Mol Cell Proteomics. 2012; 54(7): 434-55.





