Interested in our services
Contact our experts to provide further information



Methylation is a crucial modification of proteins and nucleic acids, regulating gene expression and silencing. It is closely linked to various diseases, such as cancer, aging, Alzheimer's disease, among others, and is a vital research focus in epigenetics. Methylation modifications are classified into DNA methylation, RNA methylation, and protein methylation, with protein methylation referring to the process of methyl transfer to the guanidine group of protein lysine or arginine with thioadenosylmethionine (SAM) as the donor, catalyzed by methyltransferase. Histone methylation is the most extensively studied type of protein methylation, primarily involved in heterochromatin formation, gene imprinting, X chromosome inactivation, and transcriptional regulation.
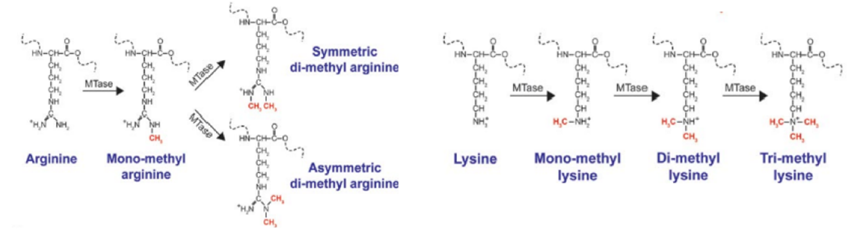
Methyltransferases play a crucial role in protein methylation and are classified into lysine methyltransferases (KMTs) and arginine methyltransferases (PRMTs). Methylation can be categorized based on the number of methyl groups added to amino acids, which can be monomethylation, dimethylation, or trimethylation. The type of methyltransferase determines whether an amino acid residue is monomethylated, dimethylated, or trimethylated. Arginine can be monomethylated or dimethylated, while lysine can be monomethylated, dimethylated or trimethylated.
PTM BIO offers protein methylation modification omics products for site identification and quantitative analysis of monomethylation, dimethylation, and trimethylation. The principle behind this method involves the enrichment of methylation-modified peptides using high-quality methylated pan antibodies, followed by loading the samples onto liquid chromatography tandem mass spectrometry for analysis. By conducting database searches and matching, hundreds of modification sites can be identified simultaneously.
1. Cell: METTL13 methylation of eEF1A increases translational output to promote tumorigenesis
This study combined with high-precision methylation modification omics found that mettl13 can be used as a lysine methyltransferase. By catalyzing the k55me2 modification of eEF1A, it can improve the GTPase activity of eEF1A, enhance the efficiency of protein translation, and promote the mechanism of cancer process. Knocking out mettl13 can significantly inhibit the development of tumors. The study revealed that mettl13, as a potential drug target, provided a new idea for the treatment of prostate cancer and lung cancer.
2. Nat Cell Biol: Arginine methylation regulates the p53 response
Through methylation modification omics, this study identified that R333, R335 and R337 of p53 can undergo methylation modification. Further research found that protein arginine methyltransferase (PRMT) 5, as a cofactor of DNA damage response co-activation complex, interacts with p53, is responsible for the methylation of p53, and affects the target gene specificity of p53. Therefore, the methylation of p53 is a potential control mechanism for its tumor suppressive function.
3. Cell Death Differ: RUNX3 methylation drives hypoxia-induced cell proliferation and antiapoptosis in early tumorigenesis
In this study, through methylation modification omics, ChIP-seq and various biochemical experiments, researchers found that methyltransferase G9a interacts with RUNX3 in hypoxic tumor cells, increasing the methylation level of RUNX3 at k129 and k171 sites, thereby reducing the acetylation of RUNX3 by P300, promoting cell proliferation and anti apoptosis, This study shows that RUNX3 may be a therapeutic or preventive target to control tumor growth in the early stage of tumorigenesis.
Interested in our services
Contact our experts to provide further information