Interested in our services
Contact our experts to provide further information



Protein phosphorylation is the process by which a protein transfers a phosphate group from ATP or GTP to a specific site on another protein, with the help of phosphorylase. The phosphorylation occurs on specific amino acid residues, such as Ser, Tyr, or Thr.
Phosphorylation is a widespread type of post-translational modification that occurs in over 30% of proteins in cells. This modification is the most basic, common, and essential mechanism for regulating and controlling protein function and viability. Phosphorylation participates in various physiological and pathological processes, such as regulating cell proliferation, development, differentiation, and apoptosis. It is widely applied in research fields related to signal transduction pathways, apoptosis, developmental differentiation, cancer mechanisms, and other life activities.
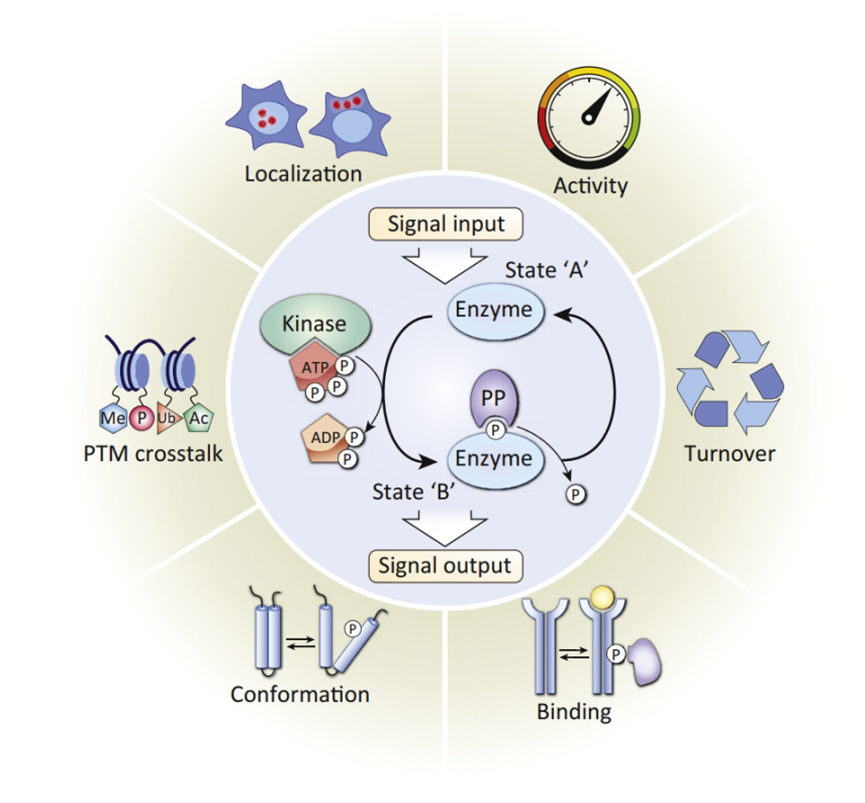
PTM BIO has launched an in-depth quantitative analysis of phosphorylation modifications by using proprietary IMAC materials for enriching of phosphorylation-modified peptides, effectively reducing sample complexity. Combined with strict double quality control and bioinformatics analysis, this powerful combination is widely used in studying the phosphorylation mechanisms under both physiological and pathological conditions.

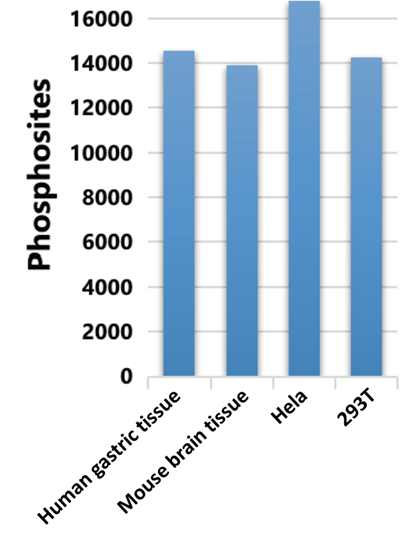
1. Technology upgrading:Identification of phosphorylation sites with deeper coverage.
Based on IMAC enrichment strategy and 4D proteomics technology, the detection sensitivity is greatly improved, and the identification number of phosphorylation modification sites is significantly increased.
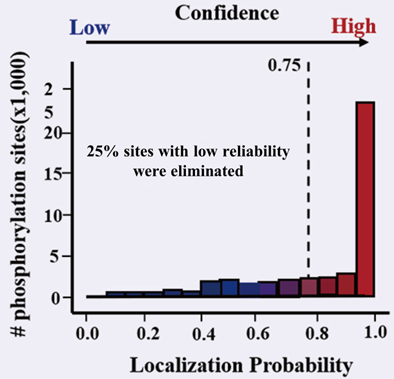
2. Quality control upgrade: Double quality control standards.
In the peptide identification of modified omics results, in addition to the card value of FDR, it is also necessary to control the FLR (false localization rate) of modified sites. Through FLR card value of 0.75, eliminate the sites with low reliability.
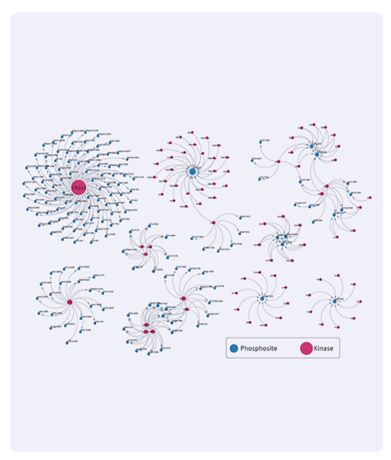
3. Bioinformatics analysis upgrade: Kinase prediction, signal pathway analysis, deep data mining.
Kinase prediction, signal pathway analysis, and in-depth data mining, are provided for exploration the mechanism under physiological and pathological conditions.
It is widely used in the research fields of signal transduction pathway, apoptosis, developmental differentiation, and cancer mechanism.
1. Signal transduction:Science: In vivo brain gpcr signaling elucidated by phosphoproteomics.
2. Cancer mechanism: Nature: Proteomics identifies therapeutic targets of early-stage hepatocellular carcinoma.
3. Immune inflammation: Immunity: Integrative proteomics and phosphoproteomics profiling reveals dynamic signaling networks and bioenergetics pathways underlying T cell activation.
4. Metabolic regulation: Cell Metab: Phosphoproteomics reveals the GSK3-PDX1 Axis as a key pathogenic signaling node in diabetic lslets.
5. Apoptosis: Autophagy: Phosphoproteome-based kinase activity profiling reveals the critical role of MAP2K2 and PLK1 in neuronal autophagy.
6. Adversity stress: Mol Cell: Reciprocal regulation of the TOR Kinase and ABA receptor balances plant growth and stress response.
7. Food science: J Agric Food Chem: Comparative quantitative phosphoproteomic analysis of the Chicken Egg during Incubation based on tandem mass tag labeling.
Interested in our services
Contact our experts to provide further information