Interested in our services
Contact our experts to provide further information




Heterogeneity renders cancer more than a single disease. This poses a significant challenge to the traditional management of cancer. Molecular subtyping of cancer, as its name suggests, is a way to classify cancers into different groups based on molecular data and classification models, it has helped researchers to identify both actionable targets for drug design as well as biomarkers for response prediction.The basis of molecular subtyping is omics data, examples include proteomics, ptms, and transcriptomics can be used for subtyping, and multi-omics data can be integrated to reveal more comprehensive and accurate conclusion. In addition, the clinical information of the corresponding patients(such as treatments, prognosis, histopathological characteristics, etc.) can be added to improve the clinical application of molecular subtyping;
Here we take NMF(Non Negative Matrix Factorization) as an example to display.
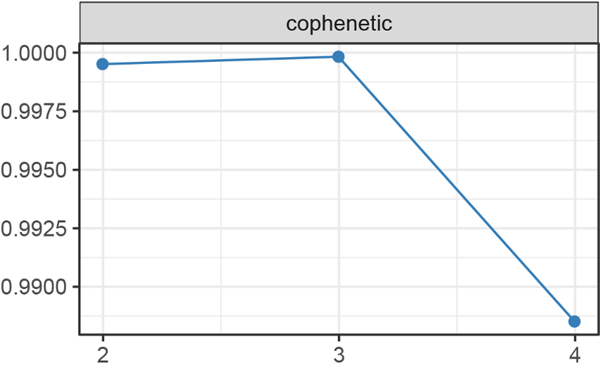
The cophenetic correlation coefficient measures the stability of the clusters obtained from NMF, Brunet proposed to take the first value of r for which the cophenetic coefficient starts decreasing.
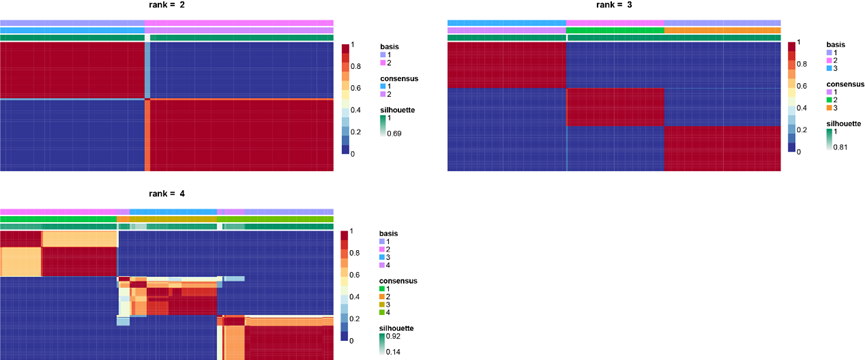
Heatmaps of the consensus matrix, for results of multiple NMF runs.
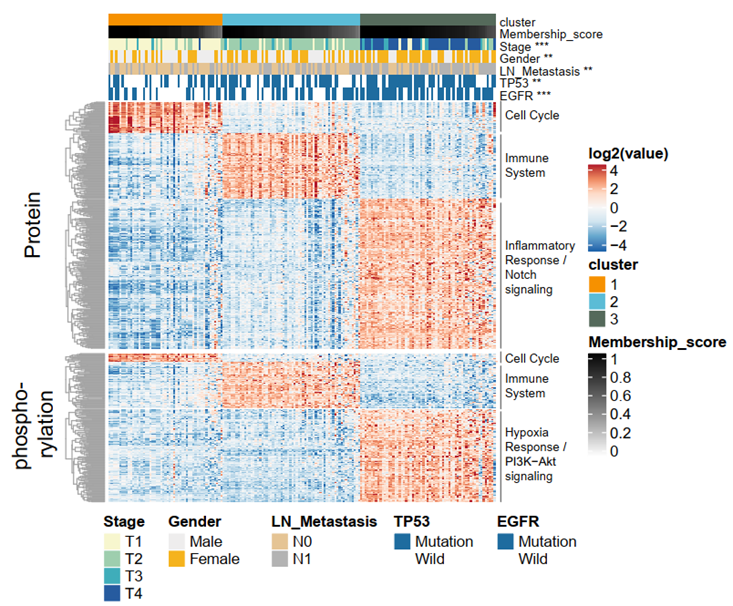
Integrative classification of tumor samples into three NMF-derived clusters and association with clinical characteristics. Within each cluster, tumors are sorted by cluster membership scores, decreasing from left to right. The heatmap shows the differential proteins and phosphoproteins for each cluster, annotated for representative pathways.
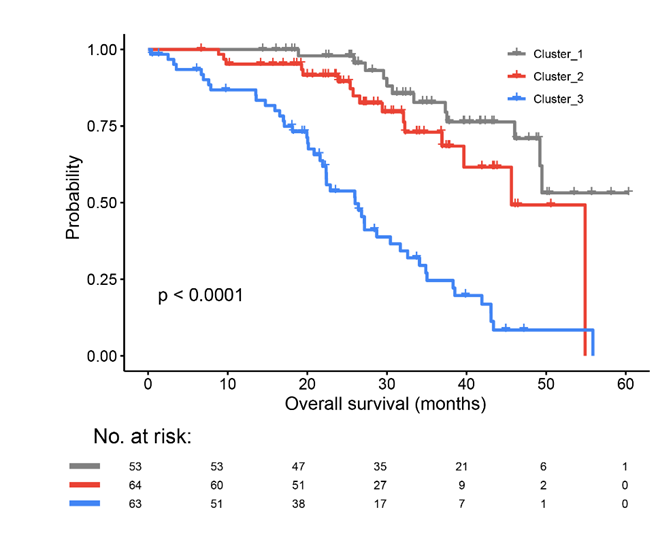
Kaplan–Meier curves of overall survival (OS) for each subtype.
Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma-2019-Nature
Interested in our services
Contact our experts to provide further information
