Interested in our services
Contact our experts to provide further information




iPath (interactive pathway explorer) is a web-based tool for the visualization and analysis of cellular pathways. Its primary map summarizes the metabolism in biological systems as annotated to date. Nodes in the map correspond to various chemical compounds and edges represent series of enzymatic reactions. It highlights general metabolic trends in multi-omics data by mapping differentially expressed proteins and metabolites.
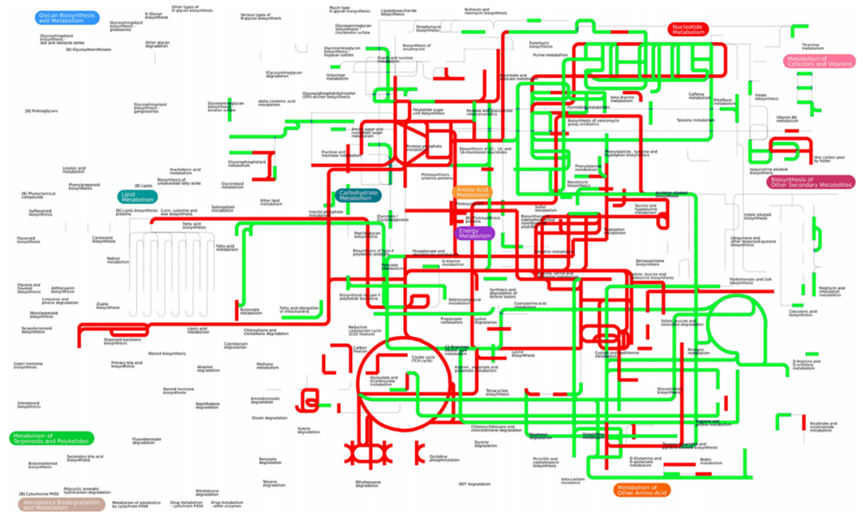
iPath Metabolic pathways annotated differentially changed compounds and reactions. In this figure, red presents up regulated reaction or compound, while green represents down regulated reaction or compound.
iPath3.0: interactive pathways explorer v3-2018-Nucleic Acids Research
Interested in our services
Contact our experts to provide further information
