Interested in our services
Contact our experts to provide further information




Ubiquitination is essential for protein degradation, transcriptional regulation and cell signal transduction in eukaryotic cells. The degree of protein ubiquitination is regulated by E1-E2-E3 cascade and deubiquitination by DUB. UbiBrowser 2.0 is a comprehensive database for collecting and predicting the interaction between ubiquitin ligase ( E3 ) and deubiquitinase ( DUB ) and substrates. We can extract the ubiquitin ligase (E3) and deubiquitinase ( DUB ) of specific Ub sites based on the database.
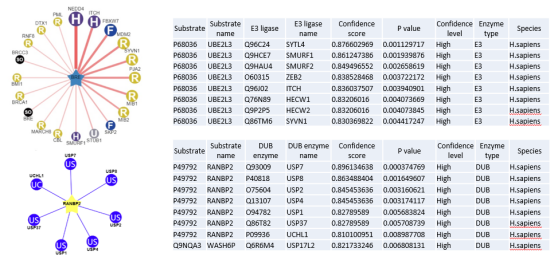
The figure shows the network diagram of BABAM2 ubiquitin ligase ( E3 ) predicted by UbiBrowser 2.0 ( upper left ), and the network diagram of RANBP2 deubiquitinating enzyme ( DUB ) ( lower left ). Text in each circle ( e.g., ' H ', ' F ' and ' SO ' ) represents the E3 ligase family.
PPDPF Promotes the Progression and acts as an Antiapoptotic Protein in Non-Small Cell Lung Cancer-2022-International Journal of Biological Sciences
UbiBrowser 2.0: a comprehensive resource for proteome-wide ubiquitin ligase/deubiquitinase-substrate interactions in eukaryotic species-2022-Nucleic Acids Research
An integrated bioinformatics platform for investigating the human E3 ubiquitin ligase-substrate interaction network-2017-Nature Communications
Interested in our services
Contact our experts to provide further information
